


Vortragsankündigung
Prof. Dr. Michael Griebel
Universität Bonn
Fast Numerical Methods for Molecular Dynamics Simulations
Abstract:
A major difficulty in MD-simulation methods is the complexity of the long
range force evaluation in each time step. To cope with this problem, there
exist various multiscale type methods, i.e. treecodes, multipole approaches
or multigrid techniques, which reduce the  complexity
of the naive approach to
complexity
of the naive approach to 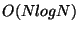 or even
or even 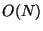 .
.
Our approach makes use of a variant of the adaptive Barnes-Hut/Multipole
method. For dealing with the adaptivity of the method, we use a hash-technique.
A further reduction of execution time is possible by parallelization. Here,
however - especially for adaptive tree-type methods - the implementation is
quite difficult and cumbersome. We present a parallel method with space-filling
Hilbert-curves. Altogether this results in an efficient parallel long-range
(e.g. Coulomb-Forces) force evaluation without potential cut-off and a simple
incooperation of short-range forces, which we have tested on several
MIMD-platforms up to 512 processors (CRAY T3E).
We will give an overview on numerical methods for molecular dynamics
simulation, discuss new reactive potenital forms which incorporate
quantum effects and present various MD-simulations with long-range
interactions with special emphasis on materials science and nanotechnology
applications.
| Zeit: | Freitag, 9. Juli 1999, 14.15 Uhr |
| Ort: | FU Berlin, Arnimallee 2-6, Raum 032 im EG |

